Research
Biobank Data Genetics Analysis
Combining genomics data and electronic health records, Biobank emerges as an important resource for precision health.
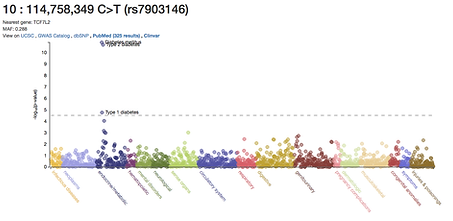
However, the size and structure of the data pose challenges to analyze them. We have developed numerous methods for GWAS and rare variant tests for biobank size data, including SAIGE and SAIGE-GENE+. These methods combine scalable mixed-effect model computation and saddlepoint approximation for accurate inference while adjusting for sample relatedness. We are also actively analyzing biobank data, including UK-Biobank and Korean Biobank (KoGES)


Genome-based Risk Prediction
With large-scale genetics/genomics data, genome-based disease risk prediction, polygenic risk score (PRS), is becoming an important tool for precision health. We are working on improving PRS, including the cross-ancestry adaptation, functional annotation incorporation, and better interpretation.
Drug effect estimation
Drug effect and side effect estimation are of great importance. By linking prescription records, genetics/genomics, and OMICs, biobanks provide an excellent opportunity to estimate drug effects and side effects at the molecular level in real-world data. We are working on causal-inference techniques and pharmaco-genomics for this purpose.




Unstructured data
Medical records include unstructured data such as medical images and clinical notes. Our research includes this domain to better utilize medical data
We are grateful to the National Research Foundation of Korea (BP+ program) and Ministry of Food and Drug Safty for supporting our research.